AI4scMed
Multiscale AI for single-cell based precision medicine

AI4scMed is a consortium (2023-2027) from the PEPR Santé Numérique (project no. 22-PESN-0002) headed by INRIA-INSERM. It gathers researchers from different institutions on AI developments for single-cell biology applied to precision medicine.
General Presentation
Cell-based precision medicine holds revolutionary potential for healthcare, but realizing its full potential demands a deep understanding of disease variability and multiscale aspects. Single-cell (sc) multi-omics offers a unique way to obtain molecular profiles of individual cells and predict disease trajectories. To harness this complexity, new AI breakthroughs are needed. Our consortium will tackle methodological challenges to bridge the gap between sc data and personalized treatments, resolving cell type differences and integrating sc-multi-omics with imaging for spatial insights.
Addressing the complexity of the human body and combining genomics with other assays, we will develop AI-based methods to handle, integrate, analyze, and visualize multiscale complexity in diseases. Our developments will leverage cutting-edge AI for sc-genomic data analysis. To infer causal mechanisms at different levels, we’ll use causal/logical/stochastic modeling to integrate heterogeneous data and account for temporal scales and biophysical priors.
We will create network inference methods to understand molecular mechanisms in clinical samples, identifying key genes and predicting therapeutic impacts. Precision medicine must also integrate variability across different cell decision levels. We aim to build predictive models, digital twins, to enable data-driven personalized treatments by connecting intracellular dynamics, biochemical processes, cell populations, and tissue-level organization.
WP1: Characterizing cellular heterogeneities
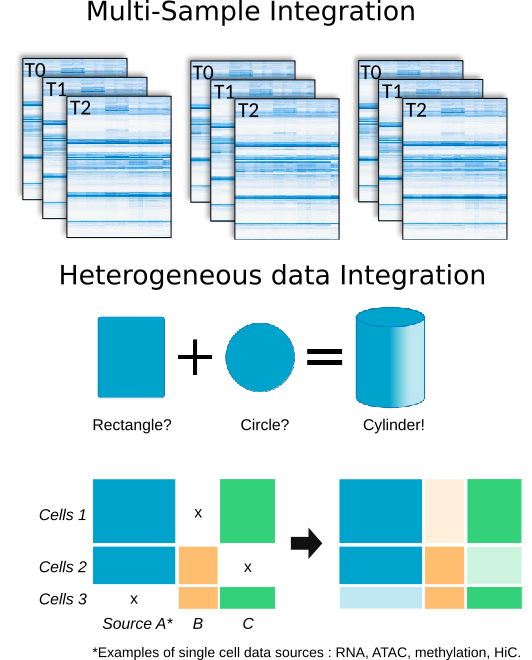
- High-dimensional sparse & stratified data,
- Joint multiple samples analysis
- Differential analysis on complex gene-expression distributions
- Integrate heterogeneous data for gene expression regulation
- Inference of gene regulatory networks
WP2: Investigating cellular ecosystems by multimodal data integration
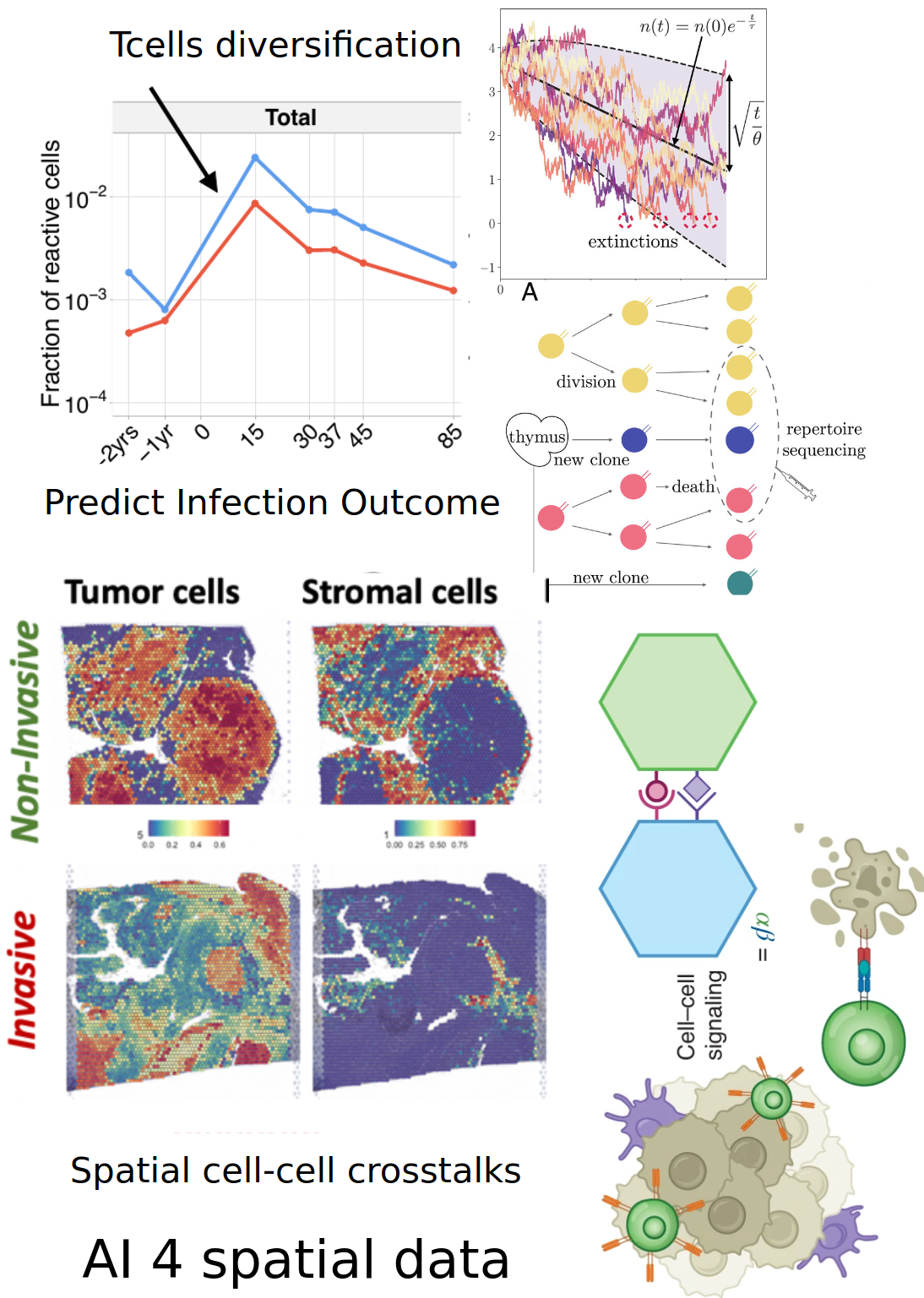
- Dynamics of immune repertoires to viral infection and cancer
- Diversification of Tcells wrt viral infection
- Tumor micro environment as an ecosystem
- Cell population communications
- Spatial machine learning & Topological
- Data Analysis
WP3: GRN inference: from dynamical to logical models
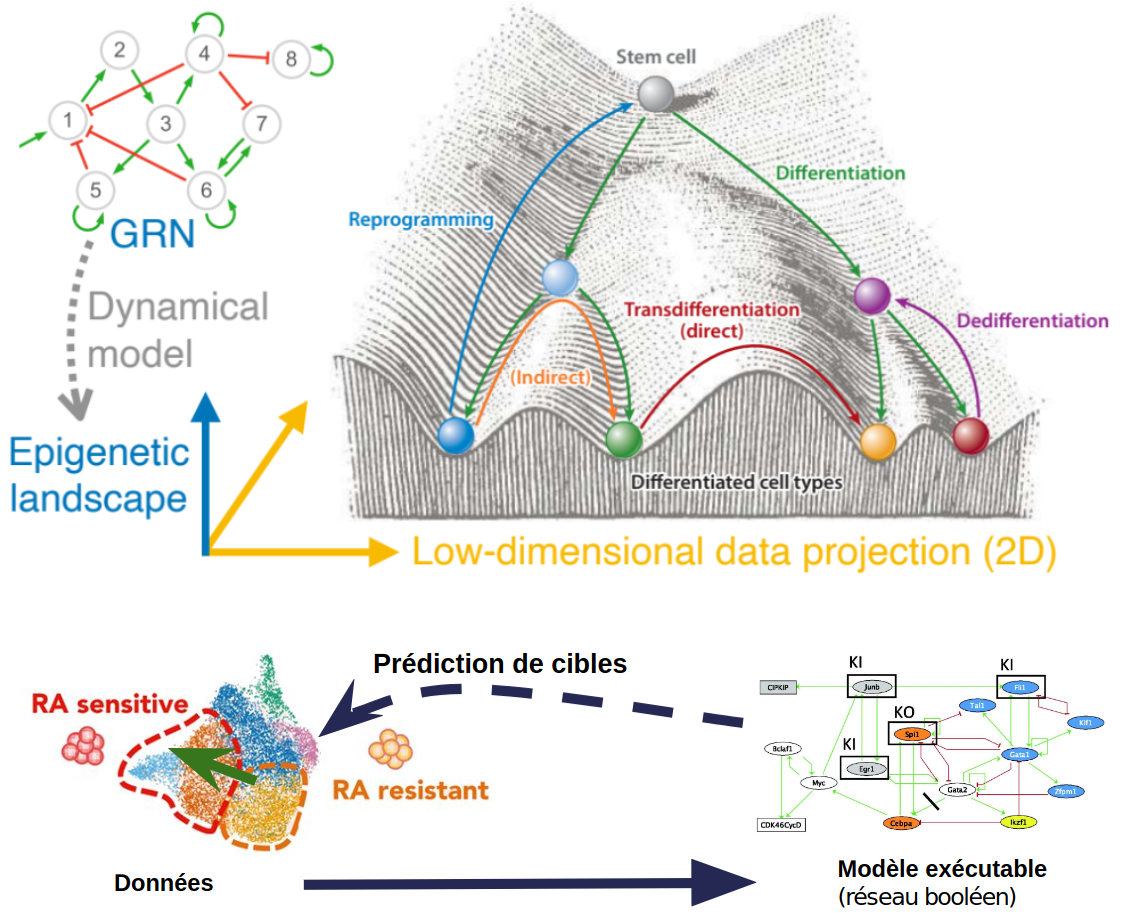
- GRN reconstruction and causal graphs from ATACSeq and scRNASeq data
- Challenge stochastic and boolean models
- Explore predicted trajectories, simulate cellular dynamics
- Predict resistance phenotype in cancer
- Predict genetic targets for treatment
WP4: Towards multiscale mechanistic models for innovative treatments
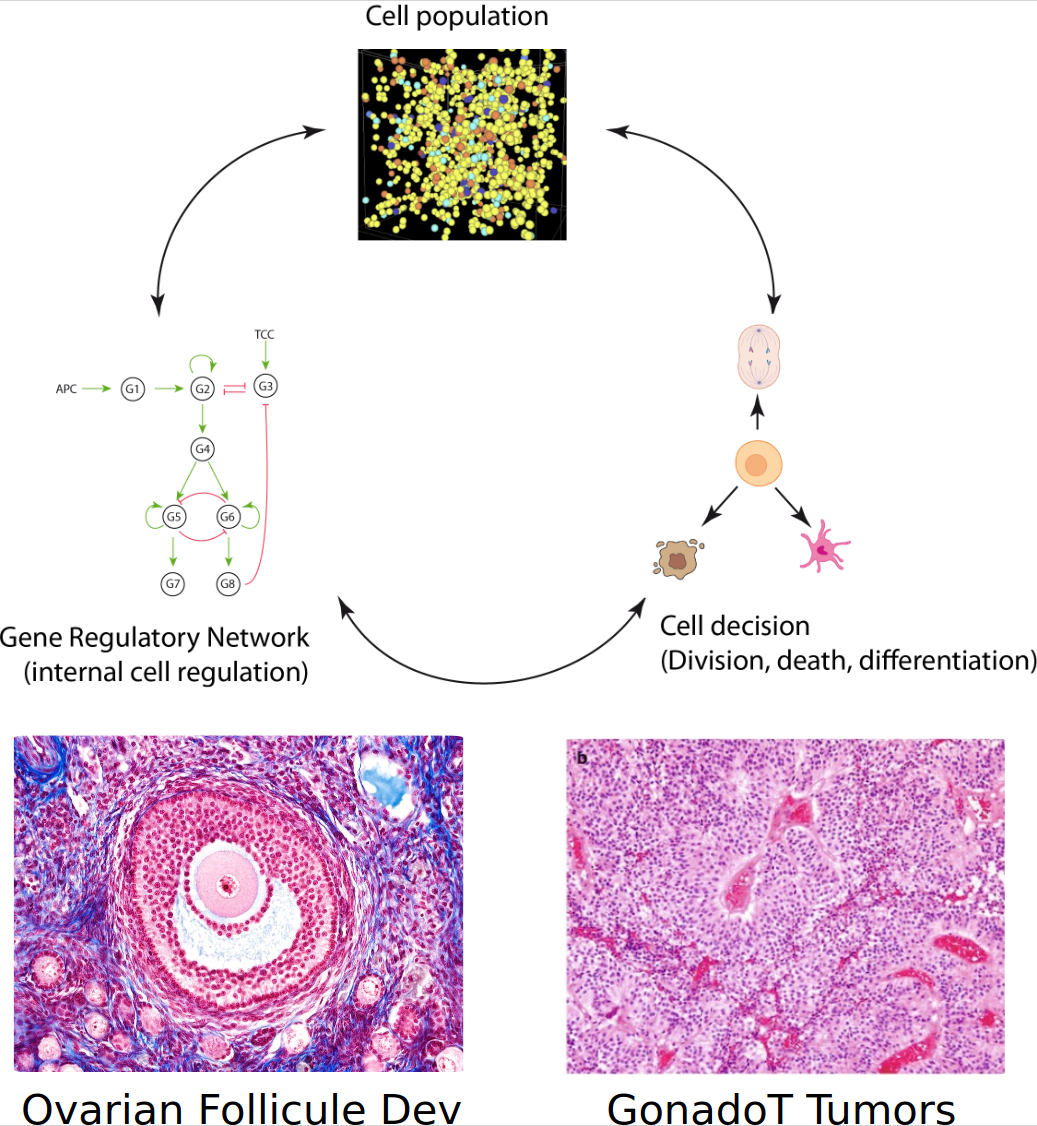
- Development of dynamical computational and mathematical multiscale models
- Interacting scales, from molecules to tissue, the cell being at the center stage
- Modeling the multiscale dynamics of spatial interactions shaping heterogeneity of gonadotroph tumors
- Investigate multiscale mechanisms underlying ovarian follicle development and their changes all along reproductive lifespan
- In silico testing of novel treatments in a patient-centered manner